|
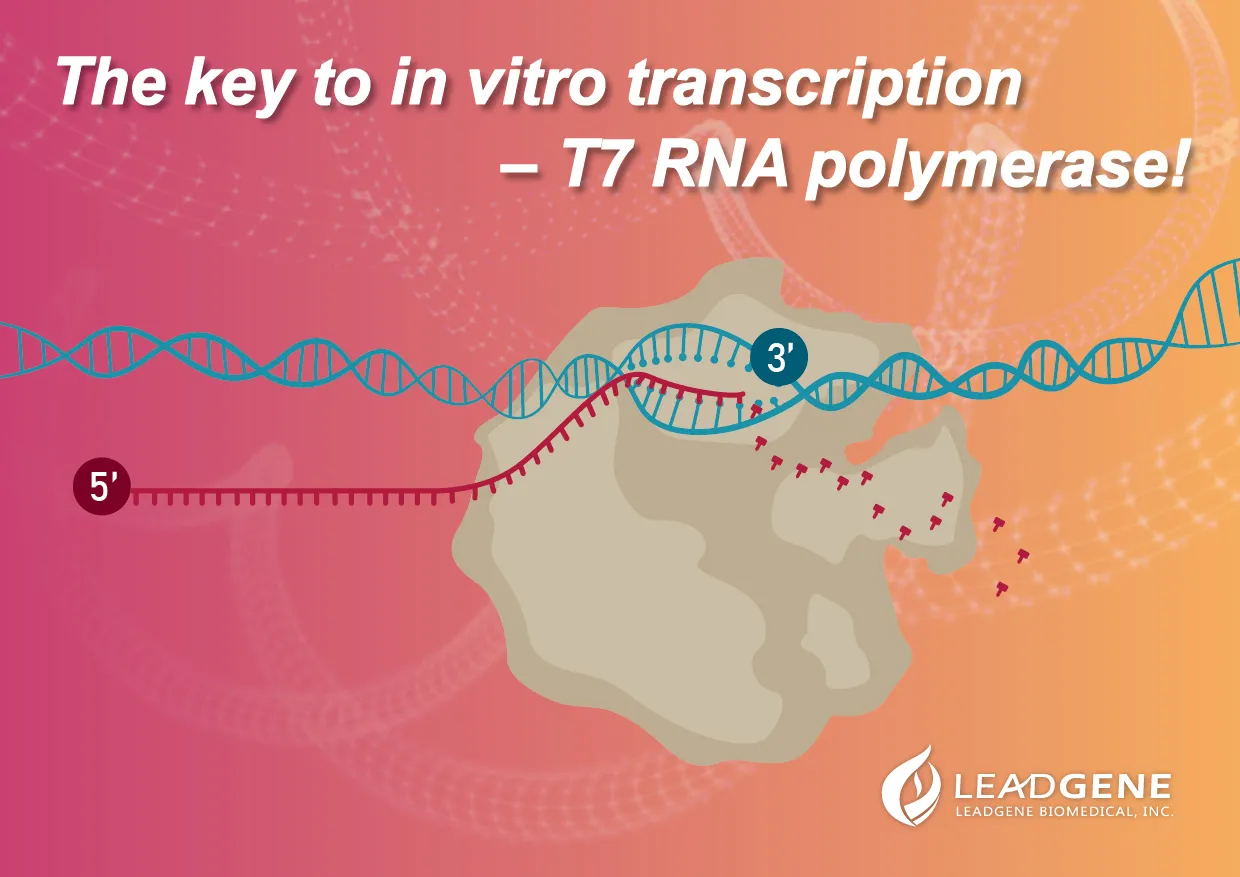
In vitro transcription, IVT |
Transcription is a process that template-directed synthesizes RNA molecules from fragmented DNAs or plasmid DNAs, and these RNA molecules encoding proteins are messenger RNAs (mRNAs). Also, there are non-coding RNAs (ncRNAs) such as microRNA, transfer RNA (tRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), or enzymatic RNA molecules (as known as ribozymes). These RNAs involve multiple functions in the body, including synthesis, regulation, and proteins processing.
In vitro transcription is the procedure that we perform transcription in a laboratory condition by combining T7 polymerases, DNA templates, NTPs, RNAase inhibitors. After the RNA synthesis, these RNAs will further proceed to 5’- capping and 3’-polyadenylation by enzymatic reactions.
|
Applications of IVT |
In vitro transcription helps scientists to develop diagnostic methods and study molecular biology. The applications of IVT are versatile, including synthesis of mRNAs for protein expression, the development of RNA vaccines, the generation of aptamers, gene editing, RNAi, and synthesis of viral RNA for infection experiments. The following is a brief introduction to the applications of IVT.
- mRNA vaccines
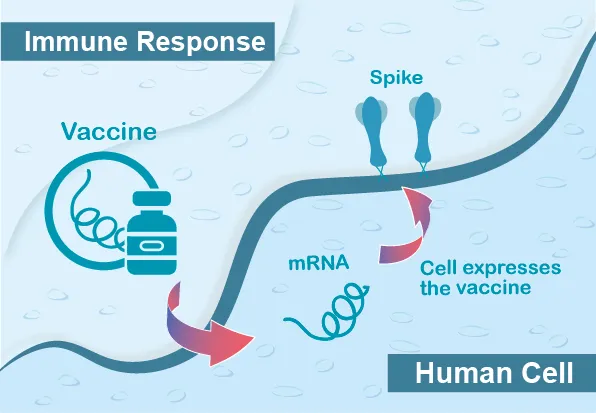
mRNA vaccine consists of lipid nanoparticles (LNPs) wrapped with messenger RNA (mRNA) encoded viral antigens. When we inoculate the mRNA vaccine, the mRNA will translate into viral proteins, and these viral antigens will further induce the immune response in our bodies.
mRNA is relatively unstable and easily degraded by RNase. Therefore, the generation of high-quality and stable RNAs is critical to the downstream application. 5’- capping, 3’- polyadenylation, and the structure of untranslated regions (UTRs) are associated with the stability of mRNAs. For a long time, the negative charge on the RNA molecules hinders the delivery of RNA into cells and becomes an obstacle for RNA vaccine development. In recent years, nanotechnology platforms have been applied to molecular biology or medical biology. The innovation of LNPs introduces the RNA vaccine to the world successfully.
Compared with traditional vaccines, mRNA vaccines are safe and efficient to produce. mRNA vaccine does not contain live pathogens and is non-infectious. Besides, since the viral culture step does not involve manufacturing, the biosafety risk is low, and the large-scale manufacture increases much faster than the traditional vaccine.
- The generation of aptamers
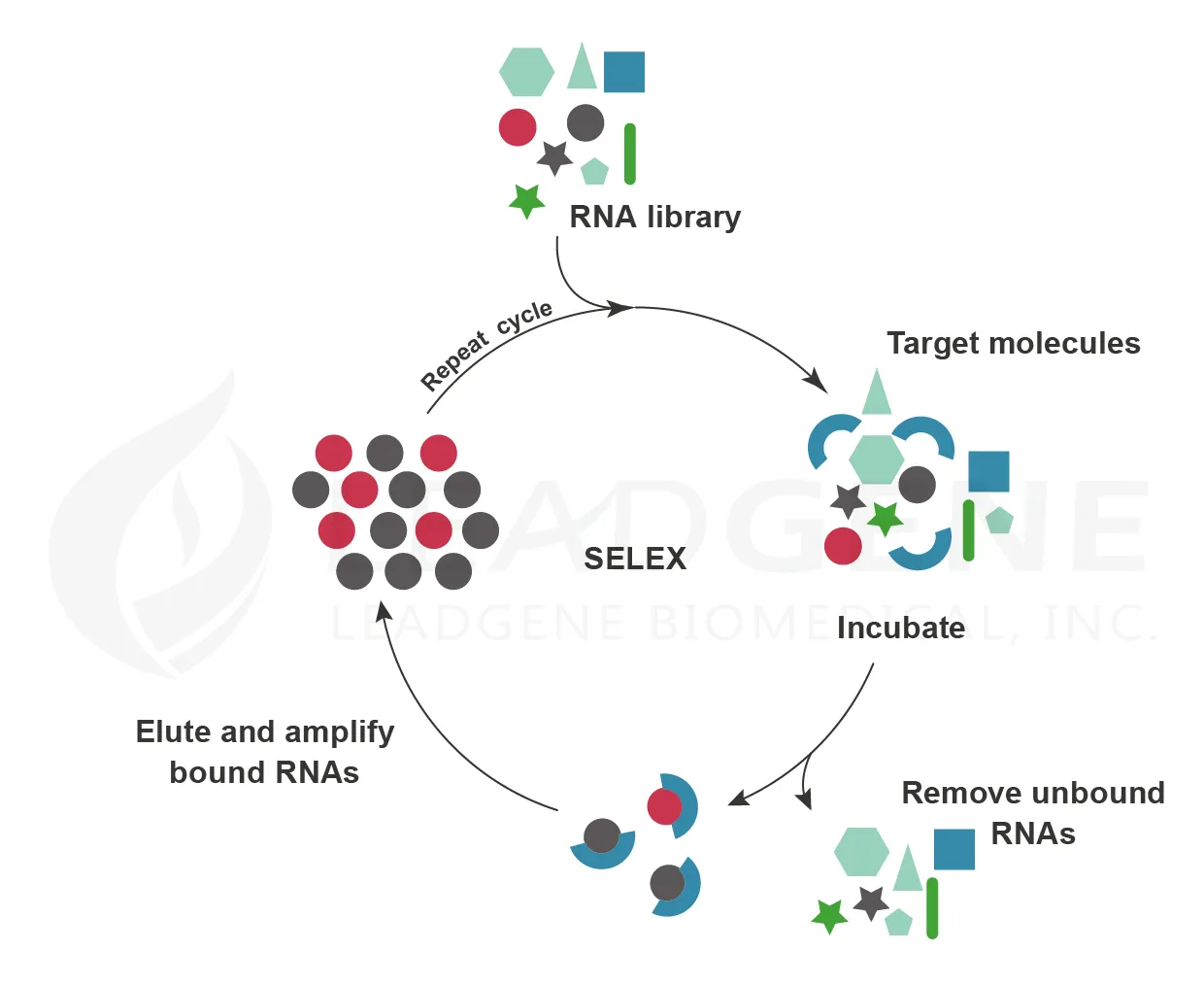
Molecular biologists Sidney Altman and Thomas Cech independently discovered that RNA participates in chemical reactions in cells. In 1992, Larry Gold and Craig Tuerk successfully selected single-stranded RNA ligands against different organic molecules by SELEX (systematic evolution of ligands by exponential enrichment). These RNAs are known as aptamers. Aptamers bind to their targets through shape complementarity and show high specificity and affinity. Moreover, aptamers can recognize several materials such as ions, small molecules, proteins, nucleic acids, viruses, bacteria, and cells. And unlike antibodies, aptamers are thermally stable and can tolerate a wide range of pH values. Therefore, aptamers are promising molecules for small molecule drugs. Macugen (Pegaptanib) is an anti-vascular endothelial growth factor (VEGF) aptamer for the treatment of neovascular age-related macular degeneration (AMD) and was obtained FDA approval in 2004. Along with the gradual development of RNA synthesis, aptamer technology shows a great potential to become therapeutic drugs.
- Gene editing
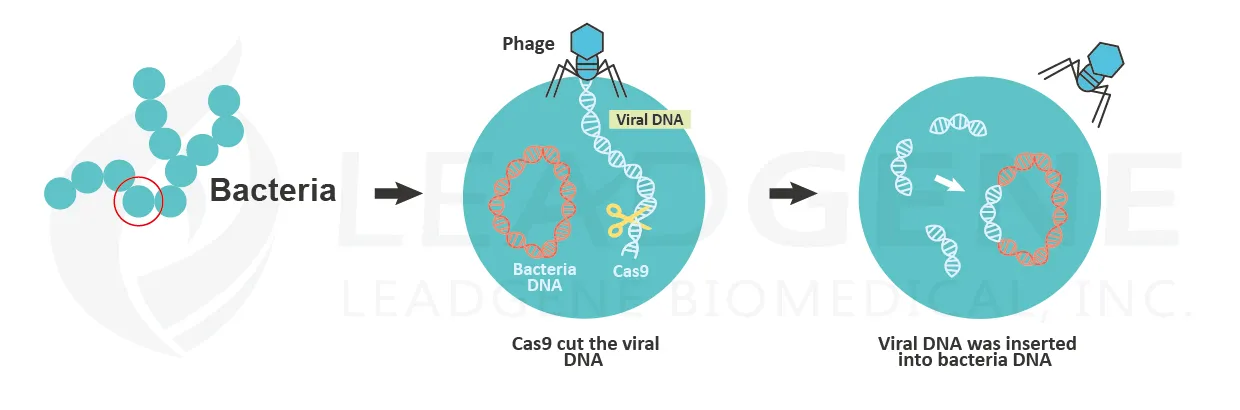
Emmanuelle Charpentier and Jennifer Doudna are the Nobel Prize in Chemistry winners in 2020. They discovered CRISPR/Cas9 genetic technology. The CRISPR/Cas9 system allows scientists to change the DNA sequences with high precision. The discovery of CRISPR dates to 1987. A Japanese scientist noted clustered DNA sequences containing palindrome in the genome of E. coli. The particular sequences were nominated as CRISPR (clustered, regularly interspaced, short palindromic repeats). After that, more and more studies showed that CRISPR could be found in 50% of bacterial genomes. The CRISPR helps bacteria protect themselves from repeated viral infections. The repeats found in CRISPR are copies of the viral genome. When the bacteria sense the phage infection, the CRISPR transcribes into transcripts, and the transcripts are then cleaved by Cas proteins to form crRNAs. These crRNAs are associated with Cas proteins to recognize foreign nucleic acids by the anti-sense characteristic of crRNAs. Once the foreign viral genome was identified, the enzymatic Cas9 protein cut the viral genome and inactivates the viruses.
CRISPR/Cas9 is a powerful tool for gene editing, but certain problems exist. Although CRISPR works perfectly in bacteria, it can cause unwanted off-target editing. Cas9 recognizes 23 nucleotides, but there are 6.5 billion base pairs in the human genome. Therefore, CRISPR may cut the non-target sequences. Moreover, the CRISPR/Cas9 mechanism is only responsible for cutting and making a DNA double-strand break (DSB). However, in the following step, chromosome repair is also critical. In eukaryotes, repairing DSBs involves two mechanisms, homology-directed repair (HDR) and nonhomologous end-joining (NHEJ). HDR is homologous recombination; hence it’s a more accurate repair pathway. HDR happens only when the cell contains a homologous piece of DNA present in the nucleus. Otherwise, the cell turns to the NHEJ pathway. Unlike HDR, the NHEJ pathway ligates the break ends without the use of a homologous template. Hence, NHEJ is an error-prone mechanism for DSB repair. Unfortunately, human cells use NHEJ as the major pathway for DSB repair. Therefore, the cutting by CRISPR/Cas9 may trigger NHEJ repair and result in unwanted deletions or insertions.
- RNAi
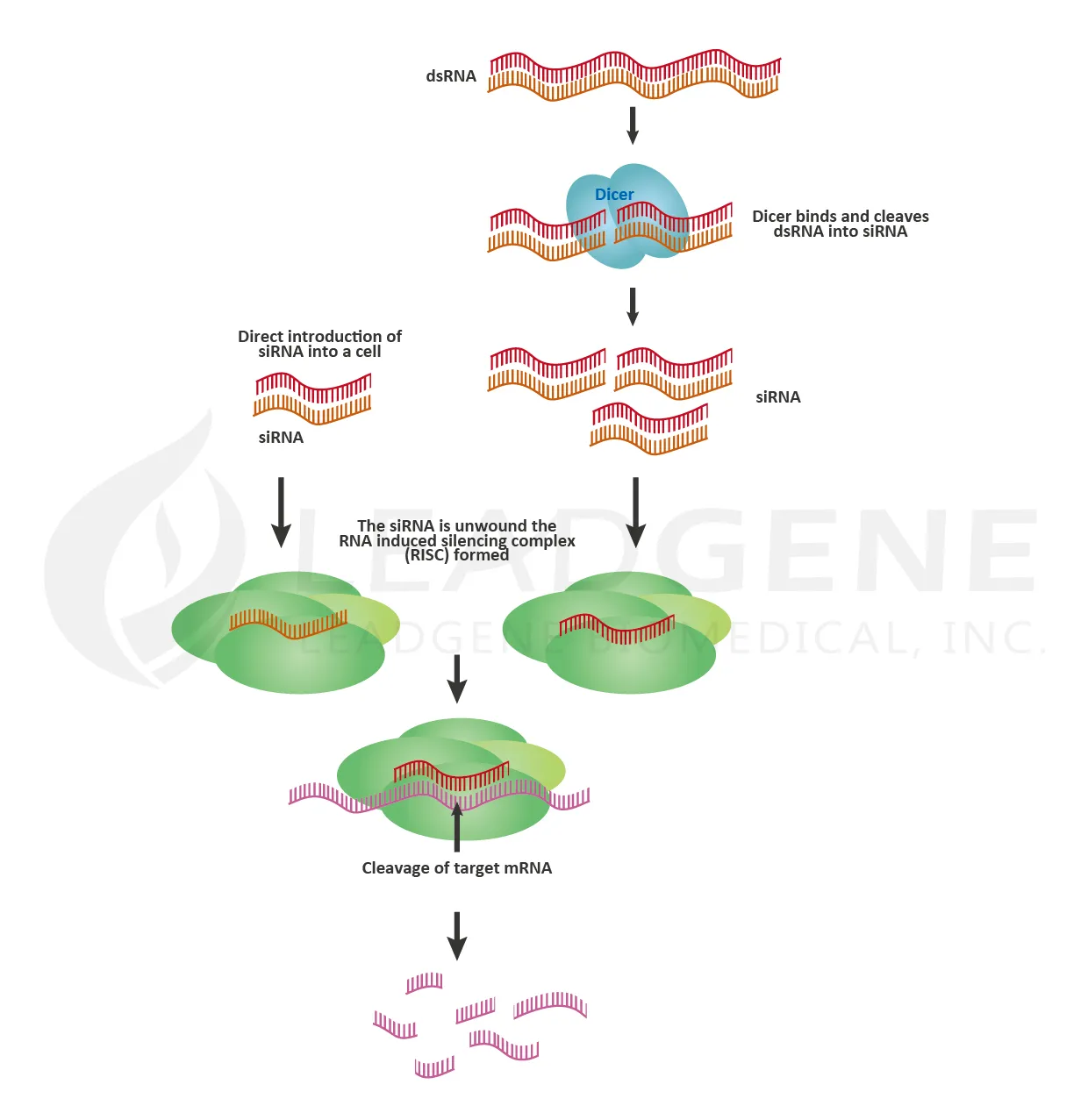
RNA interference (RNAi) is the specific suppression of gene expression by RNA molecules, and it is an efficient tool for targeted gene silencing. RNAi is a powerful research tool for therapeutic drug discovery, signaling transduction research, cellular process study, and biotechnology. The RNAi process involves microRNA (miRNA) or small interfering RNA (siRNA), which are separated into single strands and incorporated into the RNA-induced silencing complex (RISC). After integration into the RISC, the sequence-specific base-pairing leads the complex to cleave the target mRNA.
The benefit of bioinformatics and nanotechnology, the lipid nanoparticles (LNPs) technology is more and more mature, and RNAi as a strategy for small molecule drug development becomes promising. Researchers can use LNPs to deliver therapeutic siRNAs into cells to trigger RNAi. In the field of drug development, small molecules or antibodies face challenges such as the complexity of the structures, high molecular mass, and low drug-target binding affinity. However, RNA drugs are based on the characteristic of base-paring, so the binding to the target is specific. Furthermore, the easy-to-change sequences make RNA drugs are suitable for precision medicine development and adapted to the evolution of pathogens. Onpattro (Patisiran) was the first RNAi-based drug approved by the FDA in 2018. It’s a treatment for polyneuropathy in adults with hereditary transthyretin-mediated amyloidosis (hATTR). Another RNAi-based drug, Givlaari (Givosiran), obtained FDA approval in 2019. It’s a treatment for acute hepatic porphyria in adults.
- Synthesis of viral RNA
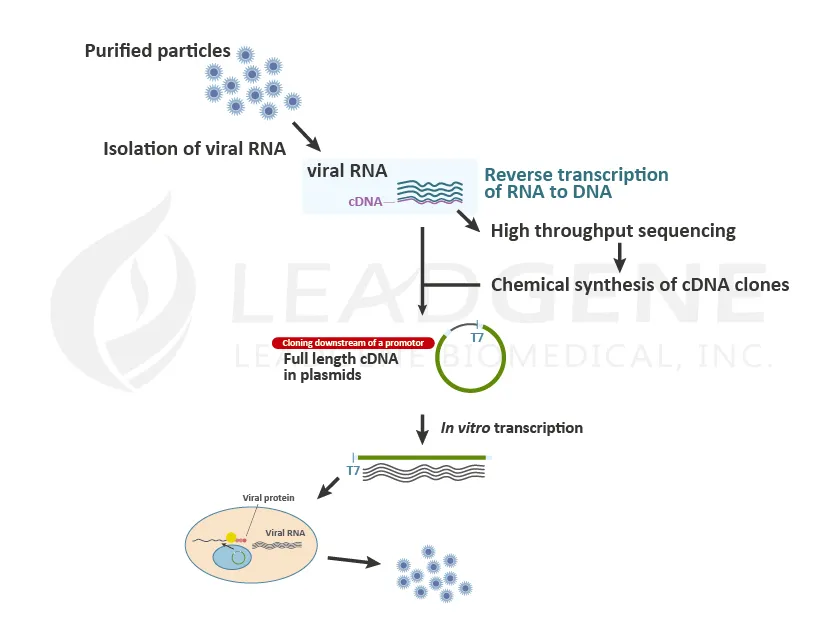
After the viruses infect and enter the host cells, the viral genome is released into the cytoplasm and starts to replicate. The viruses will suppress cellular protein expression and stat to replicate and translate the viral RNAs in the cells. Researchers generate viral RNAs by in vitro transcription and transfect viral RNAs into cells to mimic the viral infection step. Virologists apply this procedure to study neutralization assay, infection study, etc. Pharmaceutical companies use this method to culture a massive amount of viruses for vaccine manufacture. Since it is easy to introduce mutations or variations into DNA sequences by cloning technique, it is convenient to generate live-attenuated or recombinant viruses.
|
Leadgene Biomedical provides stable and high yield T7 RNA polymerase to help you accelerate your experiment! |
